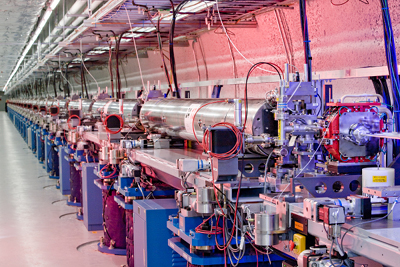
LCLS, the world's first hard
X-ray laser, is leading a revolution in coherent
X-ray imaging.
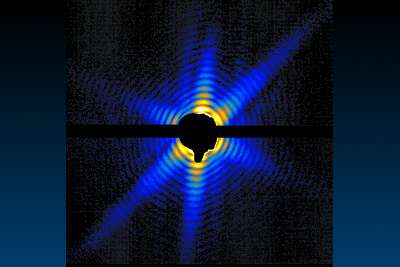
First ultrafast diffraction image of a
Mimivirus, paving the way for new methods to image
life.
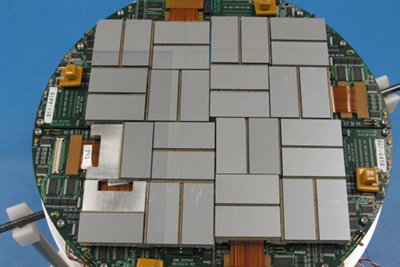
Fast X-ray detectors generate vast
amounts of data such as the CXI detector at LCLS,
capable of recording 40TB a day.
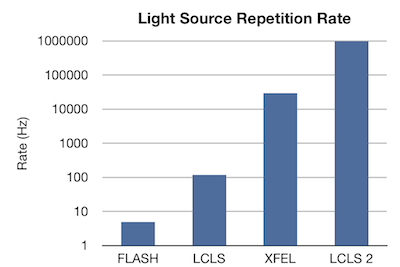
A strong data analysis
infrastructure is critical to keep up with the
exponential growth rate of X-ray free-electron lasers.
Welcome to the Coherent X-ray Imaging Data Bank (CXIDB), a new
database which offers scientists from all over the world a
unique opportunity to access data from Coherent X-ray
Imaging (CXI) experiments.
• New light sources and detectors have enabled novel experiments producing terabytes of
data per day.
• It's the dawn of the data deluge era for coherent X-ray
imaging.
• To best make use of all these data it is necessary to
make them accessible.
Accessibility is crucial not only to make
efficient use of experimental facilities,
but also to
improve the reproducibility of results and enable new
research based on previous experiments.
We must all accept that science is data and data are
science, and thus provide for [...] much improved data curation.—
B. Hanson, A. Sugden, B. Alberts
Science
CXIDB is dedicated to further the goal of making data from Coherent X-ray Imaging (CXI) experiments available to all, as well as archiving it.
The website also serves as the reference for the CXI file format, in which most of the experimental data on the database is stored in.